Coordination of cell growth and division
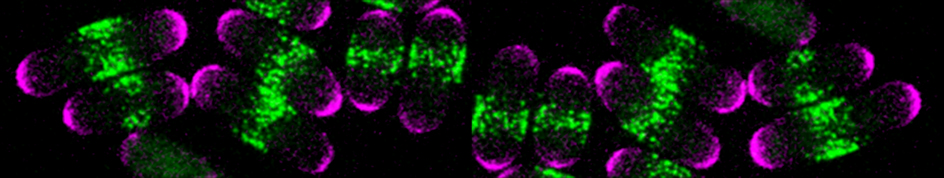
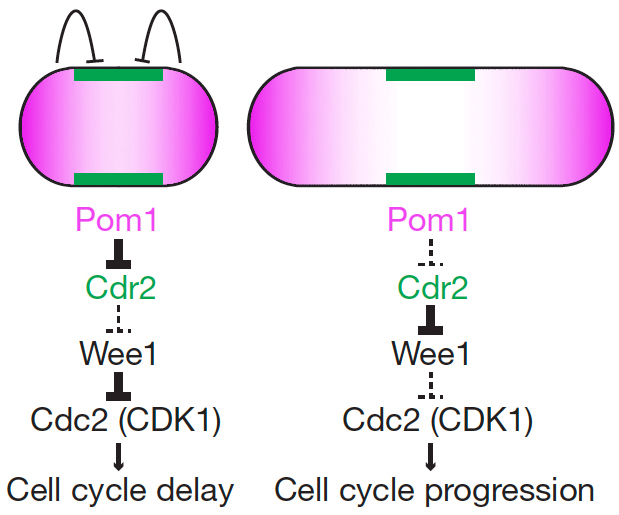
Cell cycle progression is monitored by checkpoints that ensure the fidelity of cell division and prevent unrestricted cell proliferation. Checkpoints also serve to couple cell size with division – a mechanism important to adapt to changing environmental conditions. While most studies on cell size homeostasis have focused on the links between size and biosynthetic activity, we have proposed a novel geometry-sensing mechanism by which fission yeast cells may couple cell length with entry into mitosis. The proposed system relies on two main components: the protein kinase Pom1 (purple), forming concentration gradients from the ends of the cells, and the protein kinase Cdr2 (green), itself an activator of mitotic entry that inhibit the Wee1 kinase the forms stable cortical nodes at the cell equator. As the shape of Pom1 gradients does not scale with cell length but is invariant in cells of increasing length, these gradients may serve to measure the length of the cell and couple this information with mitotic entry by inhibiting Cdr2 only in short cells.
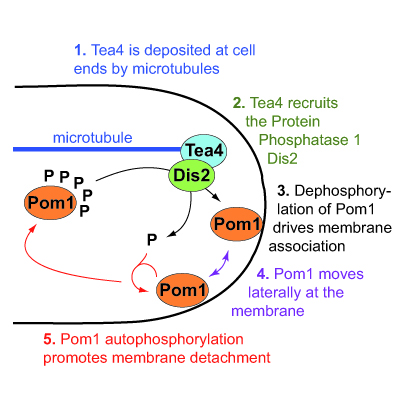
We have elucidated the molecular mechanism by which the gradients of Pom1 kinase form from cell poles. Pom1 binds the plasma membrane through a positively charged region, in a phospho-regulated manner. When unphosphorylated, this region binds the membrane with high affinity; autophosphorylation lowers membrane affinity. Gradient nucleation occurs by local dephosphorylation of Pom1 at cell poles. This dephosphorylation depends on a type I phosphatase complex (Tea4-Dis2), whose regulatory subunit Tea4 is deposited at cell poles by microtubules. Pom1 dephosphorylation drives Pom1 membrane association at cell poles, from where Pom1 likely diffuses at the plasma membrane and undergoes autophosphorylation reaction to drive membrane detachment. Through quantitative and super-resolution imaging approaches, we refined this model showing the importance of Pom1 clusters in shaping the gradient.