Expansion Microscopy developments
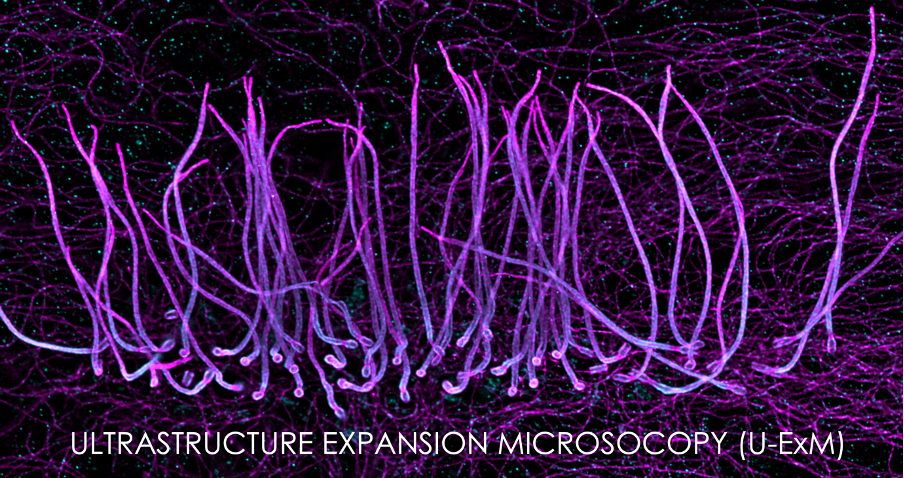
Expansion microscopy was developed in 2015 by Fei Chen, Paul W. Tillberg, and Ed Boyden (DOI: 10.1126/science.1260088). Since its invention, many variations have been made to improve the technique such as the Magnified Analysis of the Proteome (MAP) approach in 2016 which allows for volumetric expansion of the proteome (DOI: 10.1038/nbt.3641). In 2019, we developed ultrastructure expansion microscopy (U-ExM), an approach derived from the MAP protocol that allows optimal and isotropic expansion of cell architecture and macromolecules such as the centriole. Below you will find explanations and links to the different approaches we have developed-
 ULTRASTRUCTURE EXPANSION MICROSCOPY (U-ExM)
ULTRASTRUCTURE EXPANSION MICROSCOPY (U-ExM)
Using the centriole as a molecular ruler, we optimized the MAP protocol to obtain an expansion factor of 4 that preserves at best the architecture of the cellular components. We tested this protocol on different organelles such as mitochondria, cytoskeleton or the phage T4.
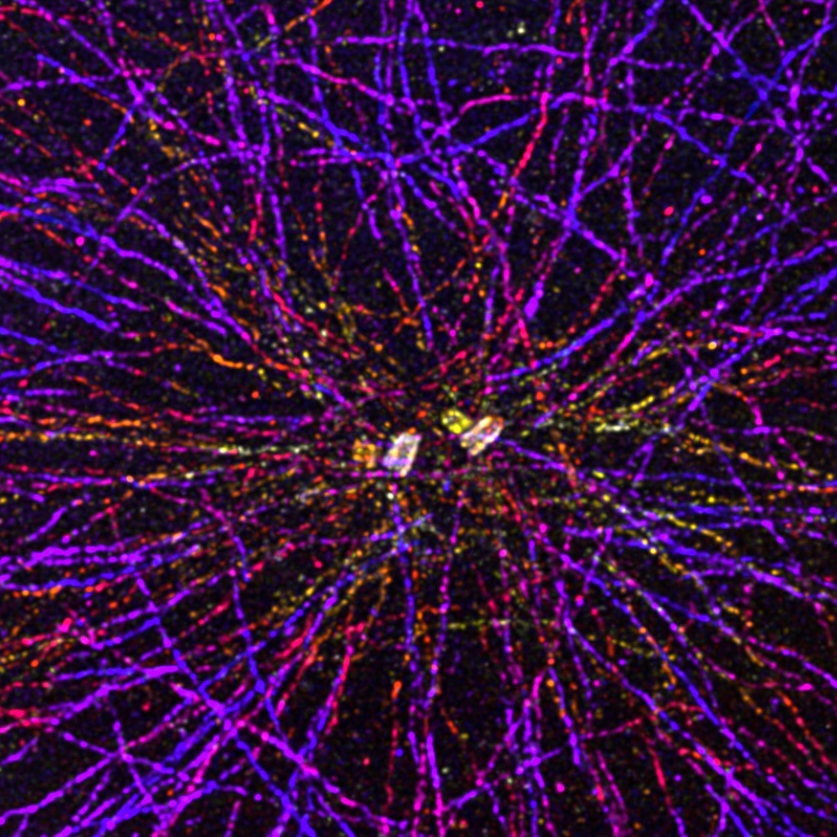
 CRYO-EXPANSION MICROSCOPY (Cryo-ExM)
CRYO-EXPANSION MICROSCOPY (Cryo-ExM)
U-ExM is a protocol developed for optimal cell expansion. However, this step requires that the cell be previously fixed chemically, which can affect the native architecture of the cell. By combining cryo-fixation, used in electron microscopy, and U-ExM, we have developed Cryo-ExM, a technique that maintains the organization of the cell before its expansion and thus allows the visualization of the cell in its quasi-native state.
Link to the article : https://www.nature.com/articles/s41592-021-01356-4
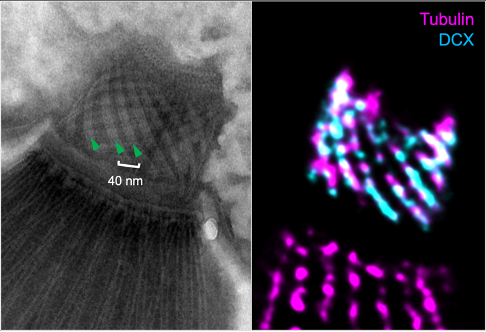 ITERATIVE U-ExM (iU-ExM)
ITERATIVE U-ExM (iU-ExM)
Expansion microscopy generally allows an expansion factor of 4. Despite numerous protocols allowing a larger expansion factor, it remains difficult to reach the same resolution than Single Molecule Light Microscopy. Here, we have developed an iterative U-ExM protocol allowing us to reach a resolution of the order of 10nm.
Link to the article : https://www.nature.com/articles/s41467-023-43582-8
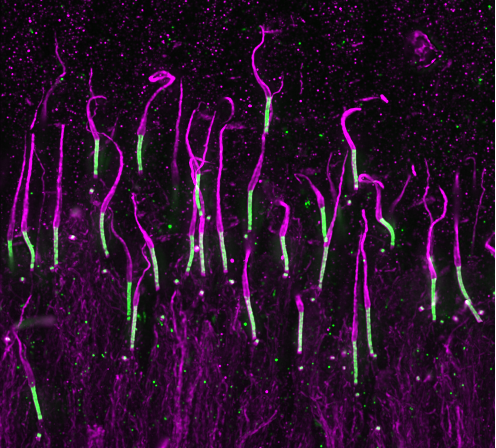 U-ExM IN TISSUE
U-ExM IN TISSUE
U-ExM works very well for soft tissue, however, as the sample is thicker than cell monolayers, some steps require longer incubations. We have tested this approach for our research on retinitis pigmentosa.
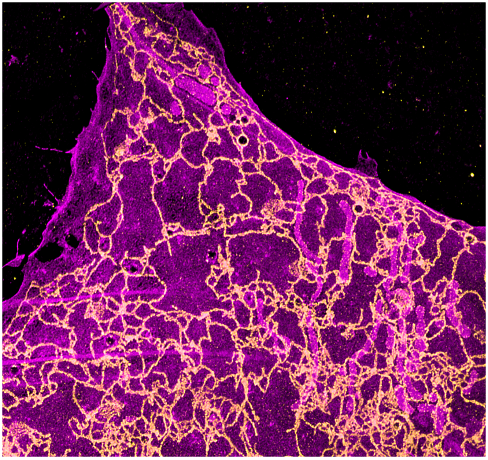
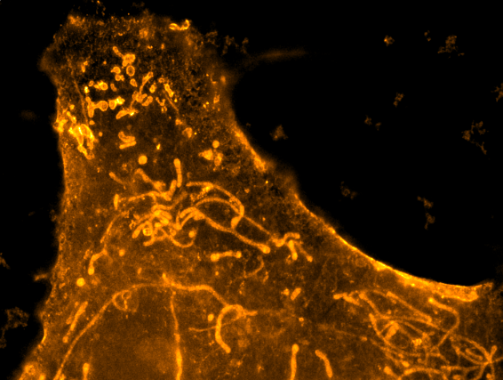 MEMBRANE STAINING in Cryo-ExM
MEMBRANE STAINING in Cryo-ExM
There are several techniques to see membranes in expansion microscopy but most of them require specific probes. Here, we have optimized the freeze substitution approach of our cryo-ExM protocol by adding some formaldehyde and paraformaldehyde in the process. Thanks to this approach, it is possible to visualize membranes simply by using the membrane marker BODIPY.
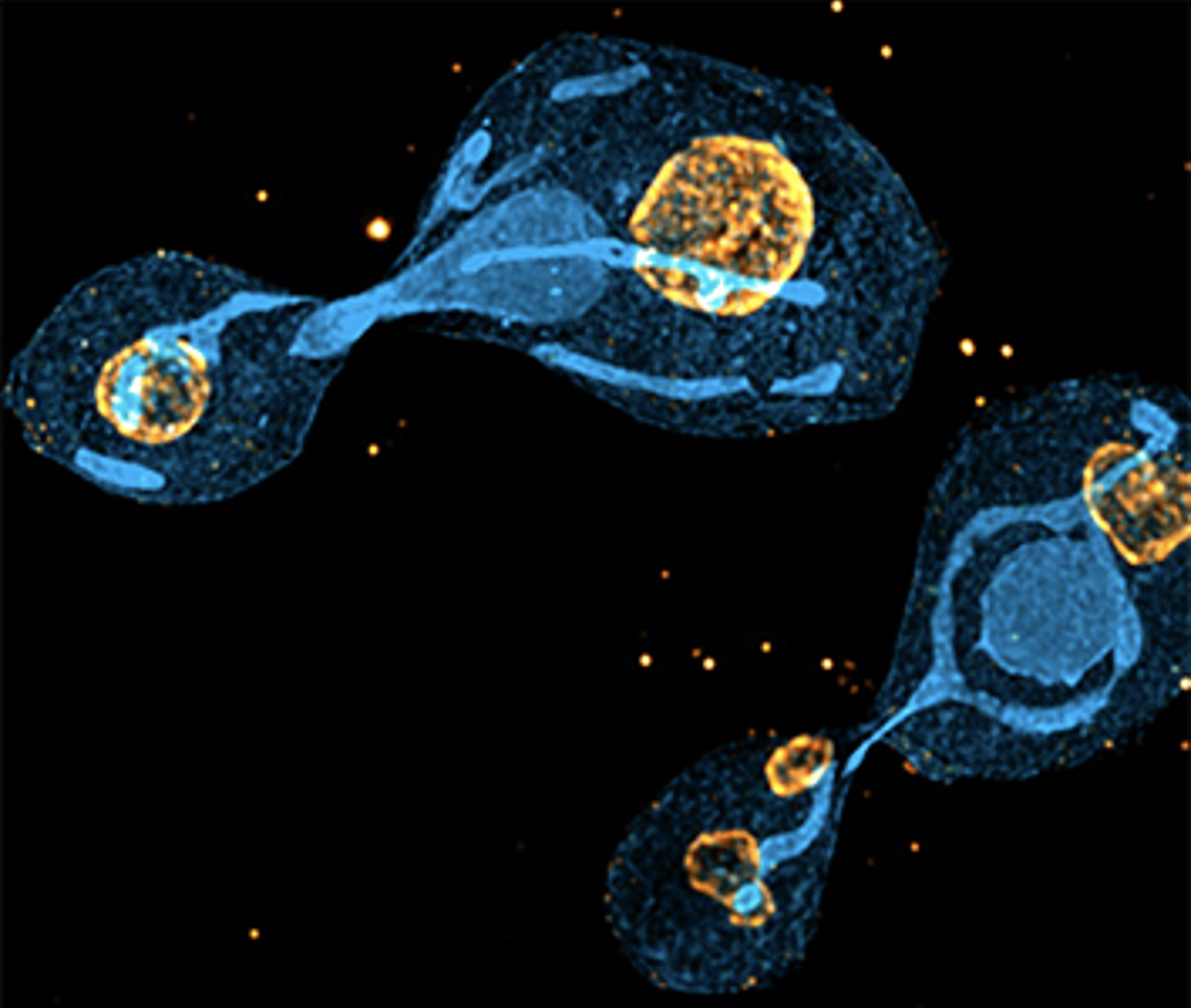 U-ExM in yeast
U-ExM in yeast
The U-ExM protocol does not require the use of proteinase K as in the original ExM protocol, making it very difficult to expand biological material with a cell wall that resists expansion. Here, by using an enzyme to digest the yeast cell wall, we have demonstrated that it is possible to use U-ExM and Cryo-ExM to visualize the architecture of S. pombe and S. cerevisiae.
You can find the article here:
 U-ExM in Chlamydomonas
U-ExM in Chlamydomonas
Chlamydomonas is a green algae that our laboratory uses to study the centriole and the asembly of the flagella. To help the community to use U-ExM, we have written a step-by-step guide. In this protocol, we also show the effects of fixations, and above all we show that cryo-EXM coupled with BODIPY staining enables us to see membrane organelles and the thylakoids.
You can find the article here :
https://bio-protocol.org/en/bpdetail?id=4792&type=0
