Publications
 An Advanced Mobile Laboratory to enable field-based microbial ecology and cell biology across scales
An Advanced Mobile Laboratory to enable field-based microbial ecology and cell biology across scales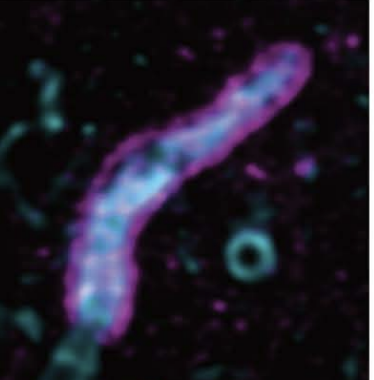
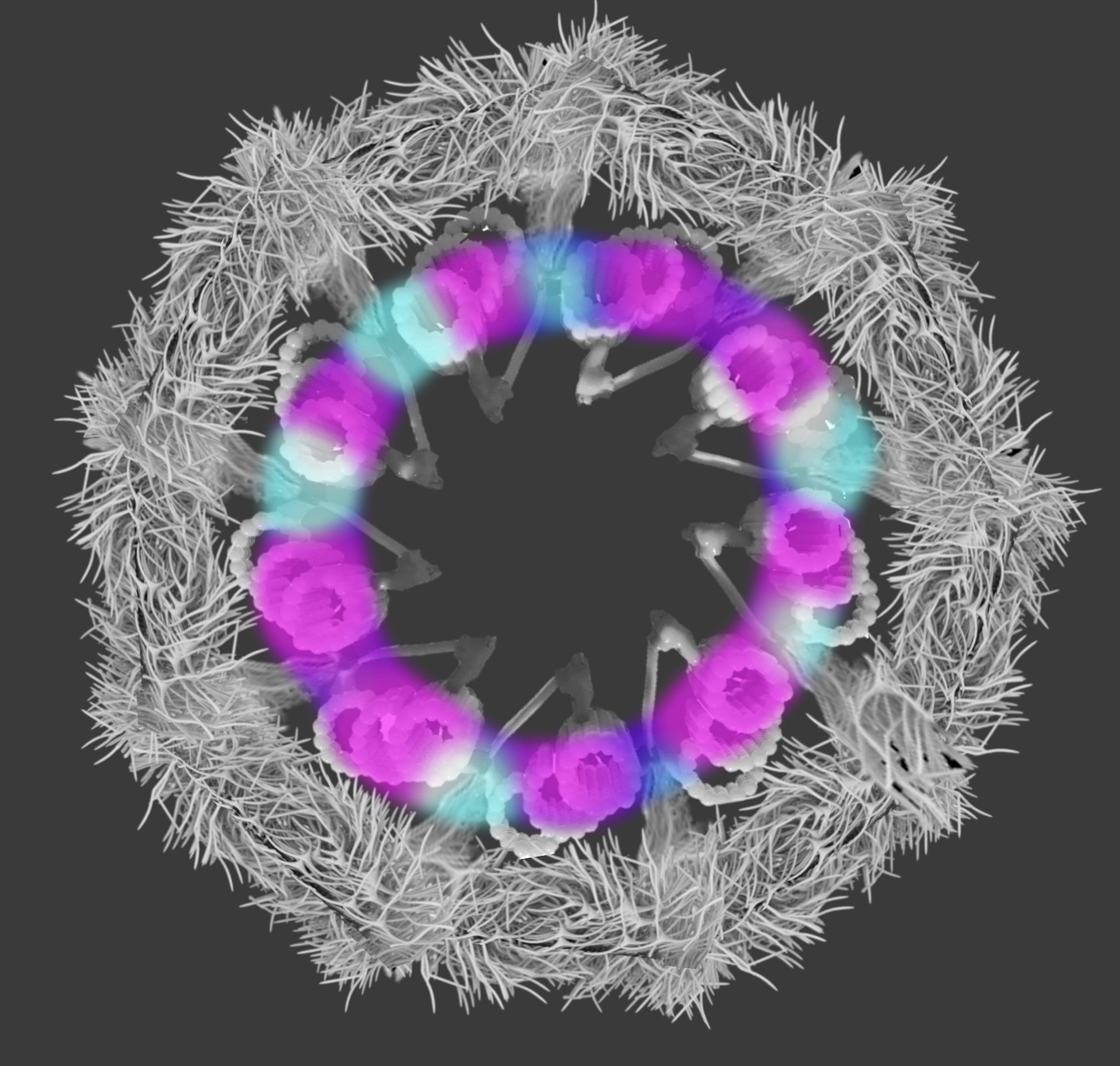

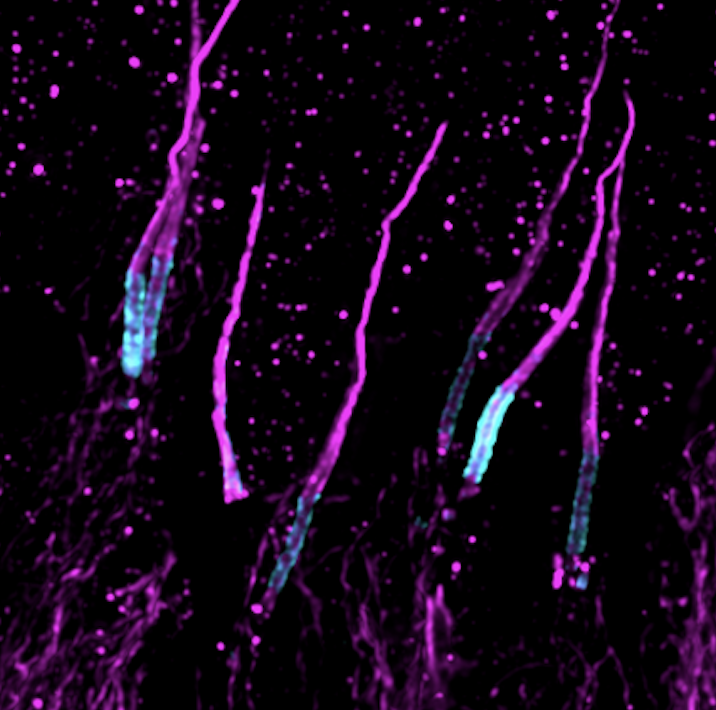
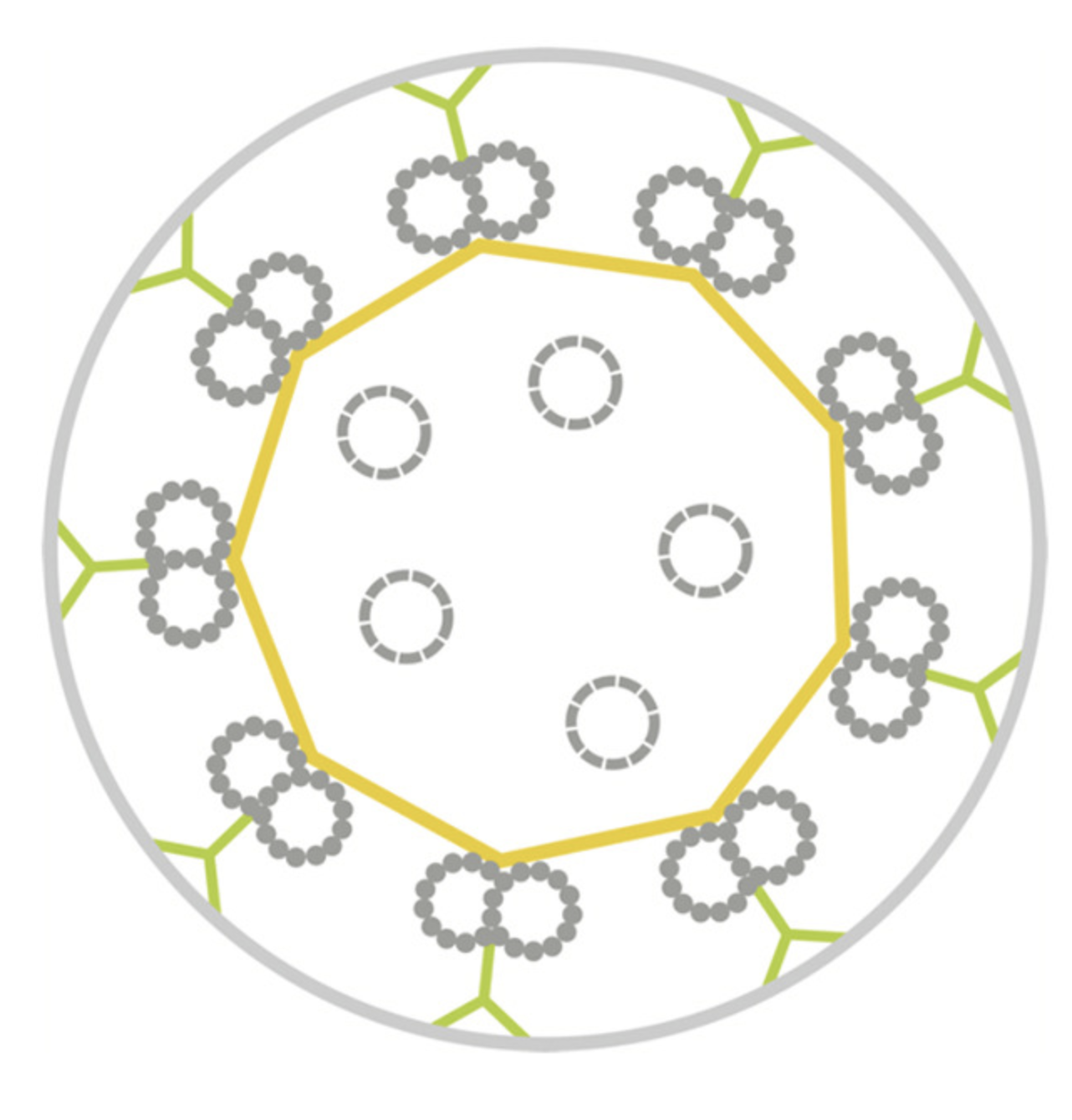




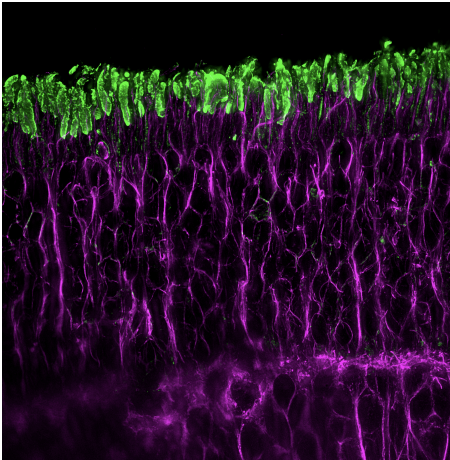 Probing the sub-cellular mechanisms of LCA5-Leber Congenital Amaurosis and associated gene therapy with expansion microscopy
Probing the sub-cellular mechanisms of LCA5-Leber Congenital Amaurosis and associated gene therapy with expansion microscopy
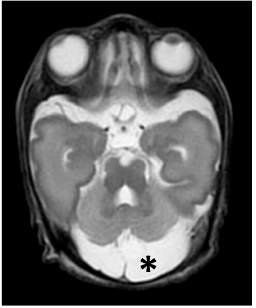 Deficiency of the minor spliceosome component U4atac snRNA secondarily results in ciliary defects in human and zebrafish. Khatri D., Putoux A., Cologne A., Kaltenbach S., Besson A., Bertiaux E., Guguin J., Fendler A., Dupont M.A., Benoit-Pilven C. , Grotto S., Ruaud L., Michot C: , Castelle M., Guët A., Guibaud L., Hamel V., Bordonné R., Leutenegger AL., Attié-Bitach T., Edery P., Mazoyer S., Delous M.
Deficiency of the minor spliceosome component U4atac snRNA secondarily results in ciliary defects in human and zebrafish. Khatri D., Putoux A., Cologne A., Kaltenbach S., Besson A., Bertiaux E., Guguin J., Fendler A., Dupont M.A., Benoit-Pilven C. , Grotto S., Ruaud L., Michot C: , Castelle M., Guët A., Guibaud L., Hamel V., Bordonné R., Leutenegger AL., Attié-Bitach T., Edery P., Mazoyer S., Delous M.

PLOS Biology - September 2022. Link

2021
Nature communications. June 2021. Link open access
PLOS Biology. Link open access - BiorXiv - Prelights
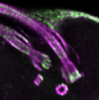 Characterization of the Novel Mitochondrial Genome Segregation Factor TAP110 in Trypanosoma brucei.
Characterization of the Novel Mitochondrial Genome Segregation Factor TAP110 in Trypanosoma brucei.
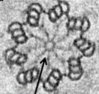
 The architecture of the centriole cartwheel-containing region revealed by cryo-electron tomography.
The architecture of the centriole cartwheel-containing region revealed by cryo-electron tomography.
Steib E., Laporte MH, Gambarotto D., Olieric N., Zheng C., Borgers S., Olieric V., Le Guennec M., Koll F., Tassin AM, Steinmetz MO, Guichard P.*, Hamel V.*
eLIFE, September 2020. Link open access - link BioRxiv
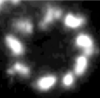 Improving the resolution of fluorescence nanoscopy using post-expansion labeling microscopy.
Improving the resolution of fluorescence nanoscopy using post-expansion labeling microscopy.
Hamel V.* and Guichard P.*
Methods in Cell Biology, August 2020. Link
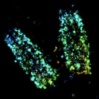 Molecular resolution imaging by post-labeling expansion single molecular localization microscopy (Ex-SMLM)., , , , ,
Molecular resolution imaging by post-labeling expansion single molecular localization microscopy (Ex-SMLM)., , , , , 
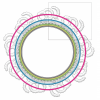 A helical inner scaffold provides a structural basis for centriole cohesion. Le Guennec M†, Klena N.†, Gambarotto D.†, Laporte MH †, Tassin AM, Van den Hoek H., Erdmann PS, Schaffer M., Kovacik L., Borgers S., Goldie KN, Stahlberg H., Bornens M., Azimzadeh J., Engel BD#, Hamel V.* and Guichard P.*
A helical inner scaffold provides a structural basis for centriole cohesion. Le Guennec M†, Klena N.†, Gambarotto D.†, Laporte MH †, Tassin AM, Van den Hoek H., Erdmann PS, Schaffer M., Kovacik L., Borgers S., Goldie KN, Stahlberg H., Bornens M., Azimzadeh J., Engel BD#, Hamel V.* and Guichard P.* 2019
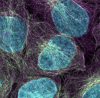 The RB530 antibody recognizes microtubules by immunofluorescence
The RB530 antibody recognizes microtubules by immunofluorescence
 Flagellar microtubule doublet assembly in vitro reveals a regulatory role of tubulin C-terminal tails. M. Schmidt-Cernohorska, I. Zhernov, E. Steib, M. Le Guennec1, R. Achek, S. Borgers, D. Demurtas, L. Mouawad, Z. Lansky, V. Hamel* and P. Guichard*.
Flagellar microtubule doublet assembly in vitro reveals a regulatory role of tubulin C-terminal tails. M. Schmidt-Cernohorska, I. Zhernov, E. Steib, M. Le Guennec1, R. Achek, S. Borgers, D. Demurtas, L. Mouawad, Z. Lansky, V. Hamel* and P. Guichard*.

2018
 Isolation, Imaging and fluorescent single particle reconstruction of Chlamydomonas centrioles. Klena1 N.§, Gambarotto D. §, Le Guennec M. , Borgers S., Guichard P.*, Hamel V.*
Isolation, Imaging and fluorescent single particle reconstruction of Chlamydomonas centrioles. Klena1 N.§, Gambarotto D. §, Le Guennec M. , Borgers S., Guichard P.*, Hamel V.*
JoVE. (139), e58109, doi:10.3791/58109. Open Access
 Reconstruction From Multiple Particles for 3D Isotropic Resolution in Fluorescence Microscopy. Fortun D, Guichard P, Hamel V, Sorzano COS, Banterle N, Gonczy P, Unser M.
Reconstruction From Multiple Particles for 3D Isotropic Resolution in Fluorescence Microscopy. Fortun D, Guichard P, Hamel V, Sorzano COS, Banterle N, Gonczy P, Unser M.
IEEE Trans Med Imaging. 2018 May;37(5):1235-1246. Open Access
 Super-resolution microscopy to decipher multi-molecular assemblies. Sieben C, Douglass KM, Guichard P, Manley S.
Super-resolution microscopy to decipher multi-molecular assemblies. Sieben C, Douglass KM, Guichard P, Manley S.
 TORC1 organized in inhibited domains (TOROIDs) regulate TORC1 activity. Prouteau M, Desfosses A, Sieben C, Bourgoint C, Lydia Mozaffari N, Demurtas D, Mitra AK, Guichard P, Manley S, Loewith R.
TORC1 organized in inhibited domains (TOROIDs) regulate TORC1 activity. Prouteau M, Desfosses A, Sieben C, Bourgoint C, Lydia Mozaffari N, Demurtas D, Mitra AK, Guichard P, Manley S, Loewith R.  Identification of Chlamydomonas Central Core Centriolar Proteins Reveals a Role for Human WDR90 in Ciliogenesis. Hamel V, Steib E, Hamelin R, Armand F, Borgers S, Flückiger I, Busso C, Olieric N, Sorzano COS, Steinmetz MO, Guichard P*, Gönczy P.*
Identification of Chlamydomonas Central Core Centriolar Proteins Reveals a Role for Human WDR90 in Ciliogenesis. Hamel V, Steib E, Hamelin R, Armand F, Borgers S, Flückiger I, Busso C, Olieric N, Sorzano COS, Steinmetz MO, Guichard P*, Gönczy P.*
 Cell-free reconstitution reveals centriole cartwheel assembly mechanisms. Guichard P, Hamel V, Le Guennec M, Banterle N, Iacovache I, Nemčíková V, Flückiger I, Goldie KN, Stahlberg H, Lévy D, Zuber B, Gönczy P.
Cell-free reconstitution reveals centriole cartwheel assembly mechanisms. Guichard P, Hamel V, Le Guennec M, Banterle N, Iacovache I, Nemčíková V, Flückiger I, Goldie KN, Stahlberg H, Lévy D, Zuber B, Gönczy P.
 The Human Centriolar Protein CEP135 Contains a Two-Stranded Coiled-Coil Domain Critical for Microtubule Binding. Kraatz S, Guichard P, Obbineni JM, Olieric N, Hatzopoulos GN, Hilbert M, Sen I, Missimer J, Gönczy P, Steinmetz MO.
The Human Centriolar Protein CEP135 Contains a Two-Stranded Coiled-Coil Domain Critical for Microtubule Binding. Kraatz S, Guichard P, Obbineni JM, Olieric N, Hatzopoulos GN, Hilbert M, Sen I, Missimer J, Gönczy P, Steinmetz MO.
 SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Hilbert M, Noga A, Frey D, Hamel V, Guichard P, Kraatz SH, Pfreundschuh M, Hosner S, Flückiger I, Jaussi R, Wieser MM, Thieltges KM, Deupi X, Müller DJ, Kammerer RA, Gönczy P, Hirono M, Steinmetz MO.
SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Hilbert M, Noga A, Frey D, Hamel V, Guichard P, Kraatz SH, Pfreundschuh M, Hosner S, Flückiger I, Jaussi R, Wieser MM, Thieltges KM, Deupi X, Müller DJ, Kammerer RA, Gönczy P, Hirono M, Steinmetz MO.
2015
 Direct visualization of dispersed lipid bicontinuous cubic phases by cryo-electron tomography. Demurtas D, Guichard P, Martiel I, Mezzenga R, Hébert C, Sagalowicz L.
Direct visualization of dispersed lipid bicontinuous cubic phases by cryo-electron tomography. Demurtas D, Guichard P, Martiel I, Mezzenga R, Hébert C, Sagalowicz L.
 Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Guichard P, Hachet V, Majubu N, Neves A, Demurtas D, Olieric N, Fluckiger I, Yamada A, Kihara K, Nishida Y, Moriya S, Steinmetz MO, Hongoh Y, Gönczy P.
Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Guichard P, Hachet V, Majubu N, Neves A, Demurtas D, Olieric N, Fluckiger I, Yamada A, Kihara K, Nishida Y, Moriya S, Steinmetz MO, Hongoh Y, Gönczy P.
 Hepatitis B subvirus particles display both a fluid bilayer membrane and a strong resistance to freeze drying: a study by solid-state NMR, light scattering, and cryo-electron microscopy/tomography. Grélard A, Guichard P, Bonnafous P, Marco S, Lambert O, Manin C, Ronzon F, Dufourc EJ.
Hepatitis B subvirus particles display both a fluid bilayer membrane and a strong resistance to freeze drying: a study by solid-state NMR, light scattering, and cryo-electron microscopy/tomography. Grélard A, Guichard P, Bonnafous P, Marco S, Lambert O, Manin C, Ronzon F, Dufourc EJ.
FASEB J. 2013 Oct;27(10):4316-26.
 Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Hilbert M, Erat MC, Hachet V, Guichard P, Blank ID, Flückiger I, Slater L, Lowe ED, Hatzopoulos GN, Steinmetz MO, Gönczy P, Vakonakis I. Proc Natl Acad Sci U S A. 2013 Jul 9;110(28):11373-8.
Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Hilbert M, Erat MC, Hachet V, Guichard P, Blank ID, Flückiger I, Slater L, Lowe ED, Hatzopoulos GN, Steinmetz MO, Gönczy P, Vakonakis I. Proc Natl Acad Sci U S A. 2013 Jul 9;110(28):11373-8.
 Use of red autofluorescence for monitoring prodiginine biosynthesis. Tenconi E, Guichard P, Motte P, Matagne A, Rigali S. J Microbiol Methods. 2013 May;93(2):138-43. doi: 10.1016/j.mimet.2013.02.012. Epub 2013 Mar 18.
Use of red autofluorescence for monitoring prodiginine biosynthesis. Tenconi E, Guichard P, Motte P, Matagne A, Rigali S. J Microbiol Methods. 2013 May;93(2):138-43. doi: 10.1016/j.mimet.2013.02.012. Epub 2013 Mar 18.
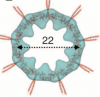 Cartwheel architecture of Trichonympha basal body. Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gönczy P.
Cartwheel architecture of Trichonympha basal body. Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gönczy P.
2011
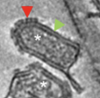 Three dimensional morphology of rabies virus studied by cryo-electron tomography. Guichard P, Krell T, Chevalier M, Vaysse C, Adam O, Ronzon F, Marco S. J Struct Biol. 2011 Oct;176(1):32-40.
Three dimensional morphology of rabies virus studied by cryo-electron tomography. Guichard P, Krell T, Chevalier M, Vaysse C, Adam O, Ronzon F, Marco S. J Struct Biol. 2011 Oct;176(1):32-40.
 Self-assembling SAS-6 multimer is a core centriole building block. Gopalakrishnan J, Guichard P, Smith AH, Schwarz H, Agard DA, Marco S, Avidor-Reiss T.
Self-assembling SAS-6 multimer is a core centriole building block. Gopalakrishnan J, Guichard P, Smith AH, Schwarz H, Agard DA, Marco S, Avidor-Reiss T.  Involvement of HFq protein in the post-transcriptional regulation of E. coli bacterial cytoskeleton and cell division proteins. Zambrano N, Guichard PP, Bi Y, Cayrol B, Marco S, Arluison V. Cell Cycle. 2009 Aug;8(15):2470-2.
Involvement of HFq protein in the post-transcriptional regulation of E. coli bacterial cytoskeleton and cell division proteins. Zambrano N, Guichard PP, Bi Y, Cayrol B, Marco S, Arluison V. Cell Cycle. 2009 Aug;8(15):2470-2.
 Visualization of proteins in intact cells with a clonable tag for electron microscopy. Diestra E, Fontana J, Guichard P, Marco S, Risco C. J Struct Biol. 2009 Mar;165(3):157-68.
Visualization of proteins in intact cells with a clonable tag for electron microscopy. Diestra E, Fontana J, Guichard P, Marco S, Risco C. J Struct Biol. 2009 Mar;165(3):157-68.







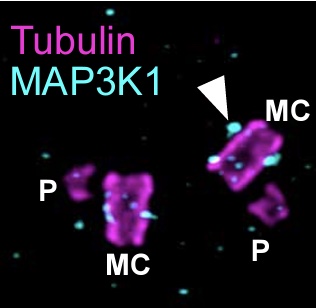
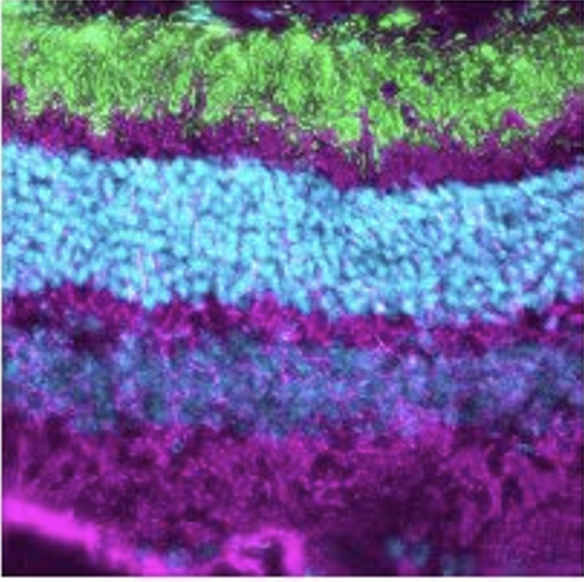


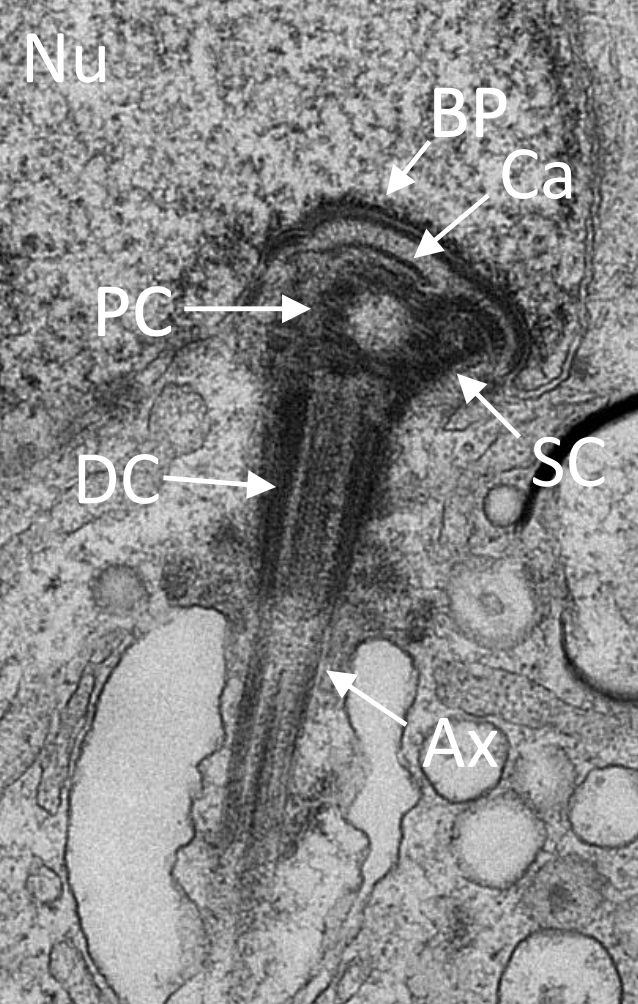


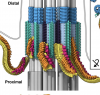 In situ architecture of the ciliary base reveals the hierarchical assembly of IFT trains.
In situ architecture of the ciliary base reveals the hierarchical assembly of IFT trains.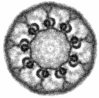

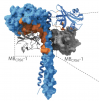 Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.