Research Projects
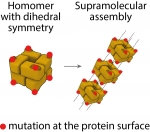
Mutations are well known for their potential to disrupt protein structure, and cells have evolved elaborate quality control mechanisms to buffer such disruption. We discovered that mutations can frequently also trigger aberrant protein assemblies. How do these assemblies impact cells? We're working on answering this question.
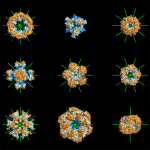
Up to 50% of proteins form homo-oligomers (homodimers, homotrimers, etc.), but this information is often unknown. We develop computational approaches to characterize this quaternary structure information with high-accuracy.

Yeast genetics techniques allow us to perturb every gene in the genome in a systematic manner, and monitor the outcome with high-throughput assays and methods. These represent a powerful way to characterize the yeast proteome's organization, function, and regulation.
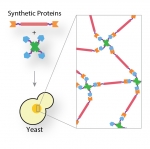
The ultimate test for our understanding of the cell machinery is our ability to alter rationally and predictably protein assemblies and pathways in cells and even to design them de novo.
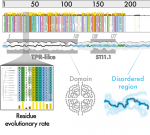
It is often easier to spot where birds live by searching for feces marks on the ground than by directly looking for birds behind tree leaves. Similarly, by searching for specific signatures in protein sequences we can reveal otherwise hidden properties of proteins, complexes, and cells.
