Research
Understanding the molecular basis of metabolic protein clusters
Enzymes frequently cluster into large higher-order-structures, also termed “metabolons”, to execute sequential, multistep cascade reactions. These self-assemblies provide several metabolic advantages, such as substrates channelling between catalytic sites, higher flux rates that are important if substrate intermediates are instable (i.e. short half-life) and they ensure a high overall catalytic efficiency of the metabolic processes.
We will use state-of-the-art microscopy methods, such as single particle analysis (SPA), correlative light and electron microscopy (CLEM) combined with FIB-SEM and time-resolved electron microscopy (TREM) to characterise the structure & architecture of such large complex assemblies. We will visualise conformational changes upon substrate binding and determine the underlying kinetics using classical biochemical and biophysical methods.
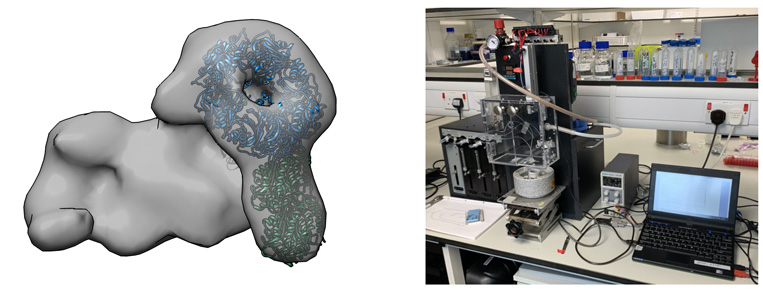
Box 1. Left panel shows the overall architecture of one of our target complexes (negative stain microscopy). Right panel shows the spraying device that will be used to conduct TREM studies.
Structural and functional studies of important cell cycle regulators
Separase is an enzyme that is responsible for cleaving the kleisin subunits (Scc1 and Rec8) of the cohesin ring that holds sister chromatids together during mitosis. Once the chromatids are liberated by separase, they segregate towards opposite poles of the cell, ready to form new nuclei in two identical daughter cells. Separase is kept in check by an inhibitory chaperone known as securin, which is intriguingly also believed to have activating properties. Although discovered almost 20 years ago, it is only recently that the structure of separase bound to securin has been elucidated.
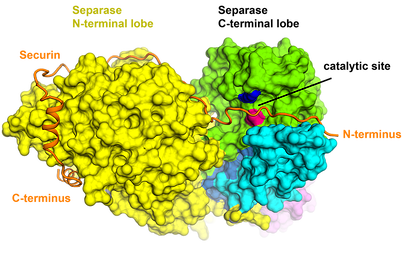
Box 2. Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Boland A et al., NSMB, 2017
It was discovered that securin forms an extended conformation to interact along the entire length of separase, and inhibits the enzyme through a pseudosubstrate mechanism at the active site. A full understanding of this interaction and nature of cell cycle control could open up new avenues for targeted drug design and will be one research branch that we will be pursuing.
Understanding the molecular basis of signal transduction
Cytokines are small soluble proteins that facilitate communication between cells in the immune and hematopoietic system. In response to external stimuli, they bind to specific cell surface receptors to trigger intracellular signalling cascades that are vital for a broad spectrum of cell functions, including proliferation and differentiation, immune responses and energy metabolism. Consequently, cytokines and their receptors are highly relevant drug targets. To elucidate the structure-function relationship of selected target receptors will be the second main branch of our lab research.
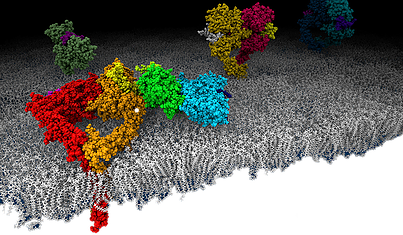
Box 3. Schematic drawing of epidermal growth factor receptor (EGFR, ErbB1, Her1) molecules on a lipid bilayer. © J. Kästner
Meet the Scientist
Past research
In animal cells, miRNAs silence the expression of mRNA targets through translational repression and/or deadenylation. Silencing requires the association of miRNAs to an Argonaute protein, which moreover binds to a member of the GW182/TNRC6C protein family. In turn, GW182 proteins interact with the two major cytoplasmic deadenylase complexes, the PAN2-PAN3 complex and the CCR4-NOT complex, to induce deadenylation and hence promote decay of miRNA targets. Structures of crucial complexes have been solved using X-ray crystallography (see References).
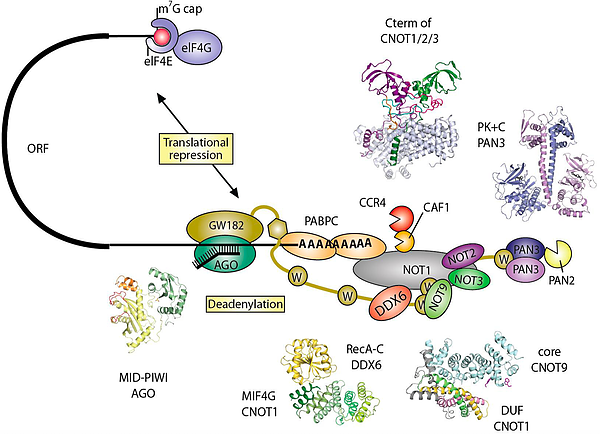
Box 4. Structural characterization of eukaryotic mRNA decay factors involved in post-transcriptional gene regulation, © Andreas Boland, PhD thesis, 2014.
