Research projects
TOR controls growth
TOR, short for Target Of Rapamycin, is an atypical serine/threonine protein kinase which curiously resembles phosphatidylinositol lipid kinases. It plays a central role in orchestrating diverse aspects of cell metabolism and physiology (Wullschleger et al, Cell 2006 ). TOR was first identified genetically in S. cerevisiae as the molecular target of the natural product antifungal rapamycin (Box 1), but is broadly conserved in nearly all eukaryotes. Inside cells, rapamycin first interacts with a proline isomerase known as FKBP12 and this complex then binds and interferes with the ability of TOR to phosphorylate a subset of substrates. In 2002, we (Loewith et al., Mol Cell 2002 ) made the important discovery that TOR proteins operate in two distinct multiprotein complexes which we named TORC1 and TORC2 (mTORC1 and mTORC2 in mammalian cells). Rapamycin-sensitive TORC1 (Box 2) and rapamycin-insensitive TORC2 (Box 3) are independently regulated downstream of environmental as well as cell-intrinsic cues. In turn, each complex independently regulates distinct aspects of biomass production and turnover necessary to maintain cellular homeostasis (Eltschinger and Loewith, Trends in Cell Biol. 2016 ).
Presently, the lab uses state-of-the-art techniques in biochemistry, structure biology, and, in collaboration with groups in a National Centre for Competence in Research (NCCR ), chemical-biology and biophysics to determine how the kinase activities of TORC1 and TORC2 are regulated downstream of signals derived from metabolites and physical properties of membranes. These efforts are supported by an in-lab Orbitrap Fusion Tribrid mass spectrometer, a new, in-house, cryo-EM platform and generous funding from the Canton of Geneva, the Swiss National Science Foundation and the European Research Council.
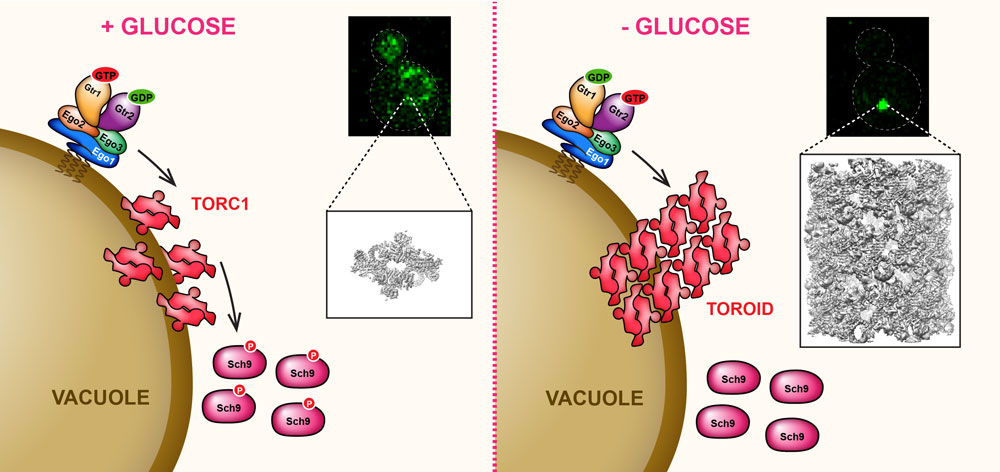
TORC1 is an ~1.2 MDa complex composed of Lst8, Kog1, Tco89 and either of the two Tor proteins (i.e. Tor1 or Tor2). TORC1 is tethered to the limiting membrane of the vacuole by the EGO – Exit from rapamycin-induced GrOwth arrest – complex (EGOC). The EGOC acts as a signaling hub in that it receives signals triggered by the presence of cellular metabolites and communicates this information to TORC1. The « business end » of the EGOC is formed by an obligate dimer of the Rag-family GTPases Gtr1 and Gtr2. Under favourable growth conditions, Gtr1 is loaded with GTP and Gtr2 with GDP and TORC1 is active; when nutrients are lacking, the guanine-nucleotide loading of the two Gtrs is reversed and TORC1 is inactive. How exactly the Gtrs regulate TORC1 activity remains an important, unsolved mystery.
In glucose-replete conditions (A in the figure), TORC1, in a Gtr1/2-dependent manner, is uniformly distributed over the surface of the vacuole. Strikingly, upon acute glucose depletion (B in the figure), TORC1 particles coalesce into a single punctum. STORM imaging suggested that this TORC1-punctum has a regular, helical structure which we confirmed by cryo-EM. Helix assembly sterically occludes the kinase active site, similarly to rapamycin-FKBP12 binding, and thus explains why the TORC1 substrate Sch9 is hypo-phosphorylated upon glucose starvation. Based on this observation we named this TORC1 structure a TOROID – TORC1 Organized into an Inhibited Domain ( Prouteau et al., Nature 2017). Presently we are trying to understand how glucose signals are sensed and, through much higher resolution cryo-EM structures, how the Rag GTPases regulate TOROID assembly/disassembly.
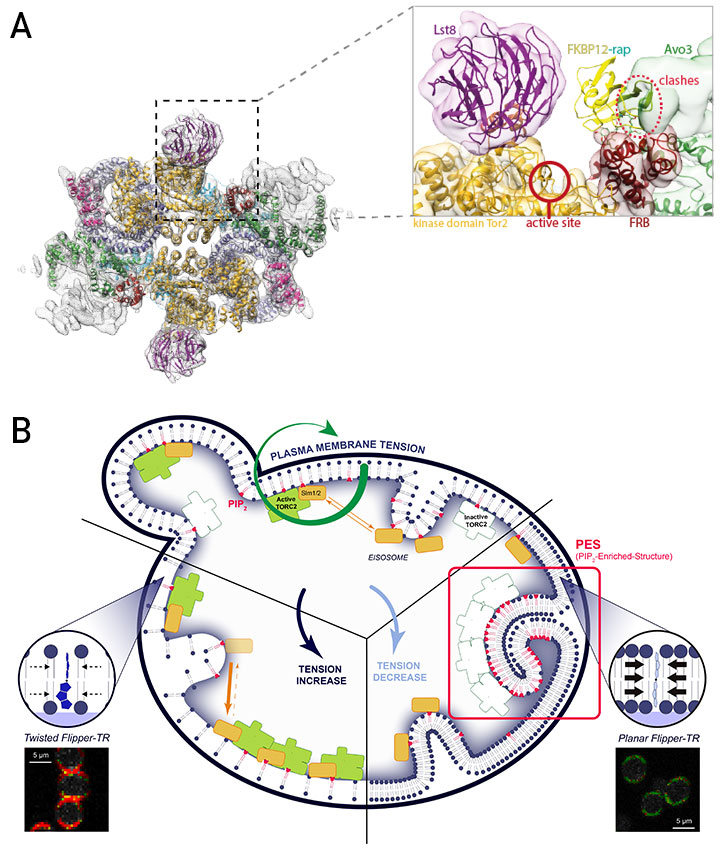
TORC2 is an ~1.4 MDa complex composed of Lst8, Avo1, Avo2, Avo3, Bit61/Bit2 and Tor2. TORC2 is inherently insensitive to rapamycin, which has made its functional characterization relatively challenging. Solving the structure of TORC2 (A in figure) revealed the molecular basis of this resistance ► the C-terminal region of Avo3 sterically occludes the FKBP12-Rapamycin binding domain of Tor2 (Gaubitz et al, Mol Cell, 2015 ; Karuppasamy et al, Nat. Comm. 2017 ). Excitingly, deletion of the C-terminus of Avo3 renders TORC2 sensitive to rapamycin and acute inhibition of TORC2 with rapamycin now greatly facilitates the study of TORC2 functions.
In parallel to these studies we discovered that TORC2 activity is enhanced by drugs that block sphingolipid biosynthesis (Berchtold et al, Nature Cell Biol. 2012 ). Building on this observation we came to realize that TORC2 is regulated, not by sphingolipids per se, but by the line tension of the plasma membrane (PM). Our association with the NCCR in Chemical Biology, particularly with the Roux, Matile and Riezman groups, helped us to develop two incredibly important tools to dissect how membrane tension regulates TORC2 (B in Figure).
The first tool was palmitoyl carnitine (PalmC), a detergent-like molecule which we identified in an ultra-highthroughput screening campaign by virtue of its ability to inhibit TORC2 signaling in vivo. The second tool was Flipper-TR®, a small-molecule probe that intercalates into the plasma membrane and, through changes in its fluorescence lifetime, reports on increases or decreases in PM tension. Using Flipper-TR we demonstrated that addition of PalmC to cells triggers a massive drop in PM tension ( Riggi et al, Nature Cell Biol. 2018). This drop in tension causes a spontaneous phase separation of PI4,5P2 into membrane domains that sequester and inactivate TORC2. Presently, we are working on our assumption that TORC2 in these domains forms an inactive super structure analogous to TORC1 TOROIDs described in Box 2. Conversely, TORC2 is hyperactivated by an increase in PM tension. This activation requires the translocation of Slm1/2 proteins from membrane invaginations known as eisosomes into domains containing TORC2 (Berchtold et al, Nature Cell Biol. 2012 ). How increased tension triggers this translocation, and how Slm1/2 binding activates TORC2 remains important questions for us to answer. Additionally, we are screening for small molecules that specifically inhibit mTORC2 with the hope that such molecules may find clinical interest to treat diseases associated with dysregulated Akt signaling (e.g. cancers, metabolic syndrome).




