Research projects
Organization AND maintenance of centromeric chromatin
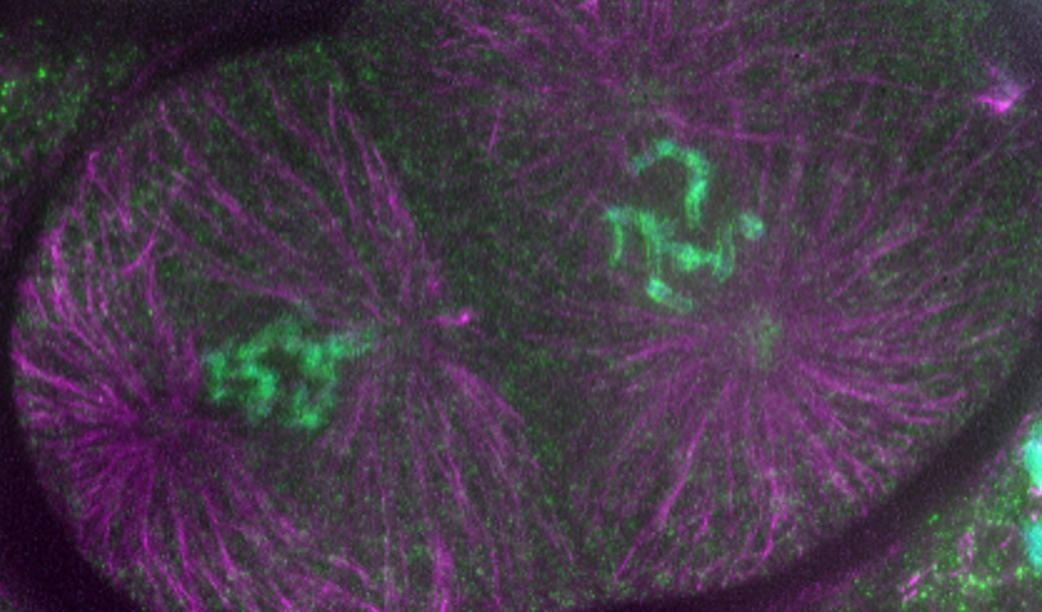
The centromere is a defining feature of eukaryotic chromosomes and is essential for the segregation of chromosomes during cell division, as it organizes the proteinaceous kinetochore for attachment to the spindle apparatus at mitosis. Aberrant centromere formation leads to aberrant chromosome segregation and aneuploidy, which is a widespread characteristic of cancer cells. In most eukaryotes, centromeres are specified by the histone variant CENP-A that epigenetically maintains centromere identity and links centromeres to the kinetochore (Schalch and Steiner, 2017). We use the nematode C. elegans to analyze how centromeric chromatin is regulated. We discovered an interplay between centromeric proteins and mitotic chromosome condensation (Wenda et al., 2021). We also identified domains of CENP-A that are crucial for setting centromere identity, but dispensable for chromosome segregation itself (Prosee et al, 2021), and we leverage these mutants to understand how centromeric chromatin is established and maintained during development, and how centromere identity is inherited from one generation to the next.
Identity and function of H3.3 in development and disease
Histone H3.3 is a replication-independent variant of histone H3 with important roles in roles in development, differentiation and fertility. The C. elegans genome contains five genes expressing H3.3. Expression of two of these genes is restricted to the germline, suggesting a role in the transmission of chromatin states at the maternal zygotic transition (Delaney et al, 2018). In contrast to other animal models, C. elegans lacking H3.3 are viable, but show embryonic replication defects and impaired activation of heat shock genes in adults (Strobino et al, 2020). The germline association and the viability of H3.3 mutant enable us to study the epigenetic contributions of this major histone variant to developmental processes.
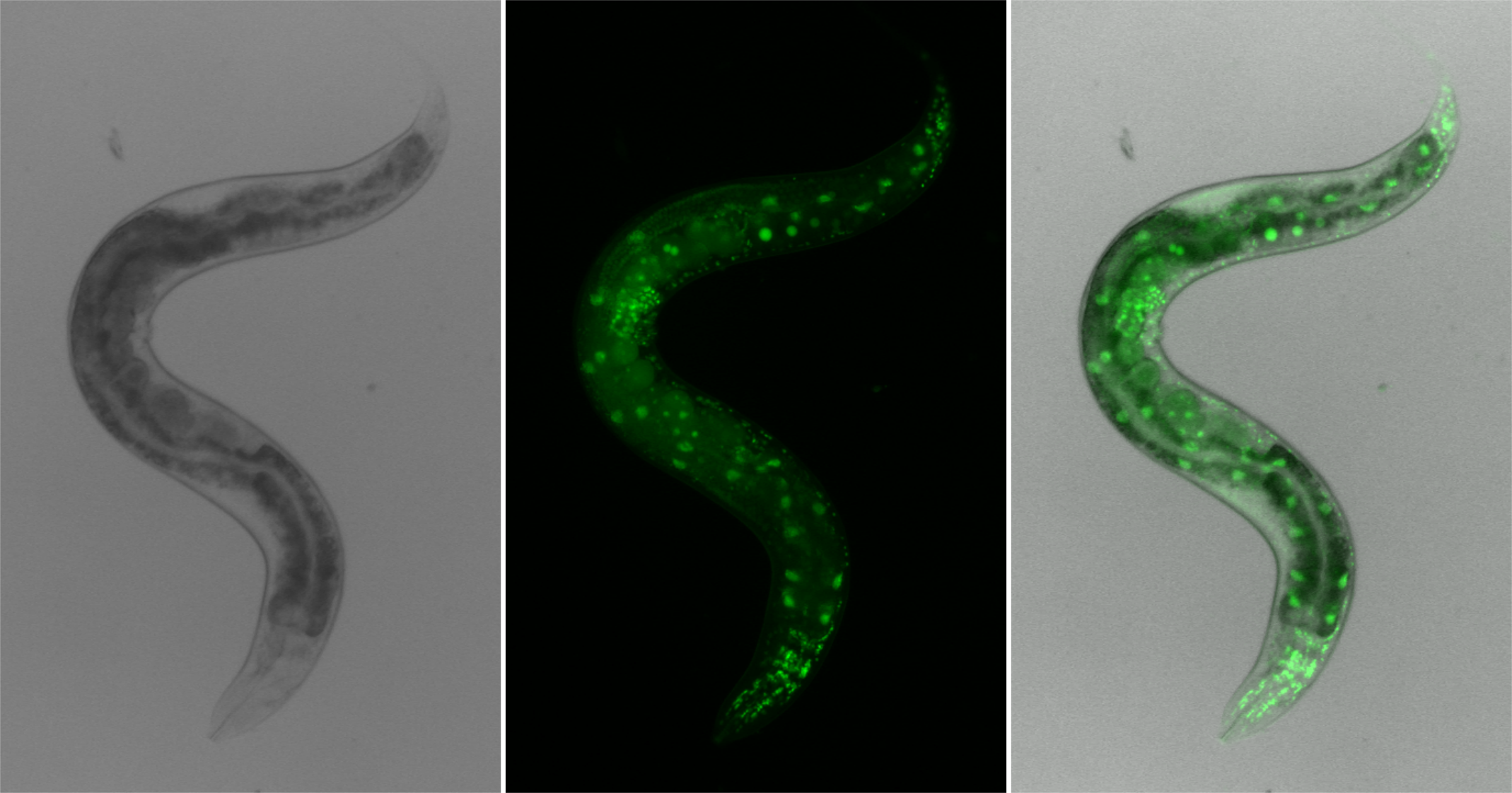
Mutation of lysine 27 to methionine in histone H3.3 has recently been discovered as a driver mutation of pediatric glioblastoma. Worms carrying the H3.3K27M mutation show ectopic activation of DNA replication and a cancer-like phenotype in the germline (Delaney et al, 2019). H3.3K27M inhibits PRC2 activity and causes a major redistribution of polycomb silencing. This leads to expression changes of hundreds of genes, but we recently showed that only specific pathways are linked to the aberrant cell cycle progression. We aim to exploit the nematode system as a model for the cancer driver mutation to elucidate how repressed chromatin is formed and inherited, and to identify potential drug targets for the treatment of this devastating disease.
